The Molecular Mechanobiology Lab
Welcome to the website for the Vanegas research group in the Department of Biochemistry and Biophysics at Oregon State University. Here, you will find relevant information about the research interests of our group, publications, news, and group members.

Research Interests
The Vanegas group uses atomistic, coarse-grained, and ab initio molecular simulation methods to understand the physical principles underlying the function of biological systems. We develop innovative state of the art molecular simulation tools in steered molecular dynamics and local stress/elasticity calculations (GROMACS-LS and MDStressLib) to address two fundamental questions in the area of nano-scale biomechanics:
1) What is the role of lipid chemical structure in the elastic properties of model biomembranes?
2) What are the activation mechanisms of mechanosensitive proteins (responsible for sensing external physical stimuli such as pressure)?

With the grateful support from

Interested in joining the group? - Looking for Ph.D. students!
News and Events
- A lot of blood sweat and tears went into this one!! Congrats to Rajitha for a great paper (highlighted under 'New and Notable' in the January issue of Biophys. J.) and thanks to our collaborators at UMD!! Read the article here ...
- Great work by undergrad Physics alumna Conner Winkeljohn and Materials Science grad student Ben Himberg for the upcoming Computational and Experimental Advances in Biomembranes Virtual Special Issue at J Phys Chem B Read the article here ...
- Hot off the press! "The mechanism of catalysis by L-asparaginase" Read more ...
- Professor Vanegas' CAREER proposal has been awarded by the NSF!! Find the project description here ...
- Lovely weather and excellent talks/posters at BPS 2020!
- Upcoming publication at Annual Reviews of Physical Chemistry "Hydration Mimicry by Membrane Ion Channels" Read the pre-print on the arXiv ...
- Our recent work "Combined molecular/continuum modeling reveals the role of friction during fast unfolding of coiled-coil proteins" is now published! Read more ...
- Our collaborative NSF MRI grant "MRI: Acquisition of a GPU Accelerated Vermont Advanced Computing Core" has been funded! Looking forward to working with UVM's new supercomputer "DeepGreen".
- Presented our work on free energy calculations of mechanosensitive channel activation at the ACS meeting in Boston (Aug. 19th-23rd, 2018)
- Our recent work "Probing key elements of teixobactin–lipid II interactions in membranes" is now published! Read more ...
- Invited seminar at Virginia Tech, Department of Physics (Apr. 30th, 2018)
- Presented our work on mechanical properties of model lipid membranes at the March APS Meeting in LA (March 5 - 9th, 2018).
- Our recent work "Ultra-thin enzymatic liquid membrane for CO2 separation and capture" is out! Read more ...
Selected Publications
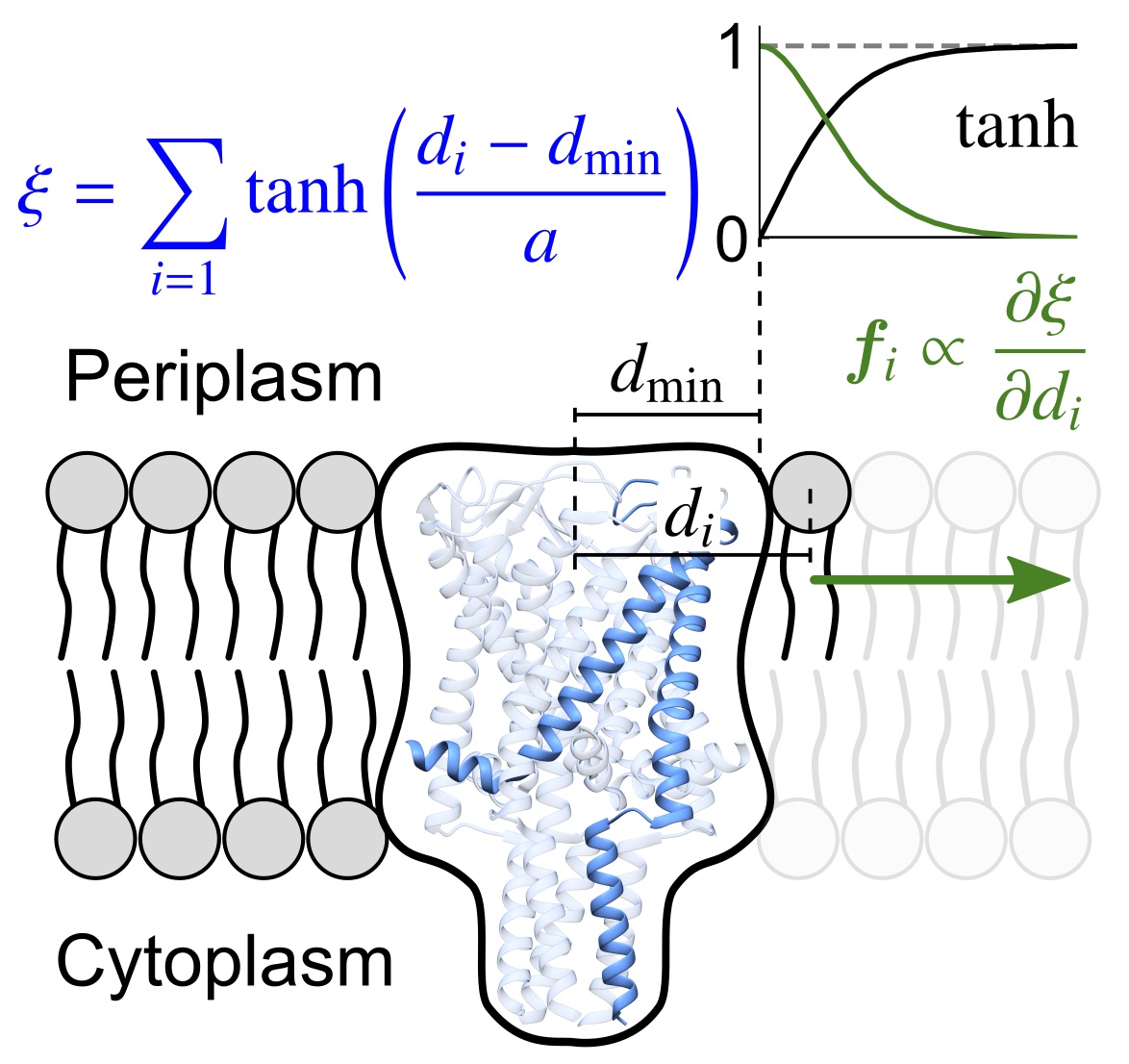 Rajeshwar T., R., Anishkin, A., Sukharev, S. and Vanegas, J.M. Mechanical Activation of MscL Revealed by a Locally Distributed Tension Molecular Dynamics Approach. Biophys. J. 19, 232-242 (2021) [PDF]
Abstract. We present a novel, to our knowledge, locally distributed tension molecular dynamics (LDT-MD) simulation method that allows application of forces continuously distributed among lipids surrounding the channel using a specially constructed collective variable. We report reproducible and reversible transitions of MscL to the open state with measured parameters of lateral expansion and conductivity that exactly satisfy experimental values.[...]
Rajeshwar T., R., Anishkin, A., Sukharev, S. and Vanegas, J.M. Mechanical Activation of MscL Revealed by a Locally Distributed Tension Molecular Dynamics Approach. Biophys. J. 19, 232-242 (2021) [PDF]
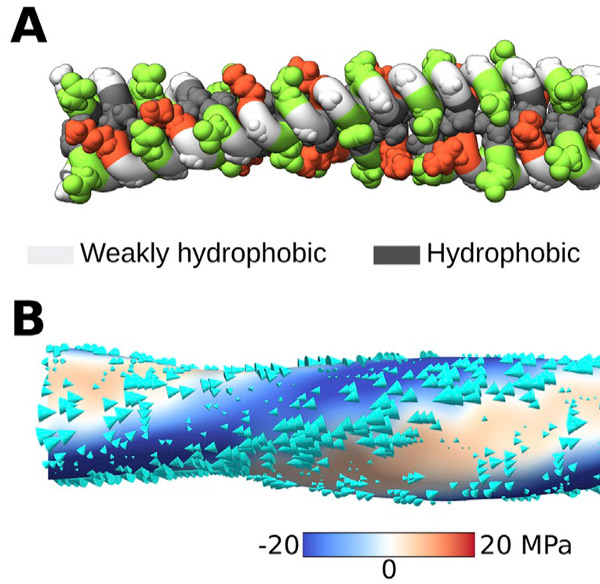
Abstract. We present a novel, to our knowledge, locally distributed tension molecular dynamics (LDT-MD) simulation method that allows application of forces continuously distributed among lipids surrounding the channel using a specially constructed collective variable. We report reproducible and reversible transitions of MscL to the open state with measured parameters of lateral expansion and conductivity that exactly satisfy experimental values.[...]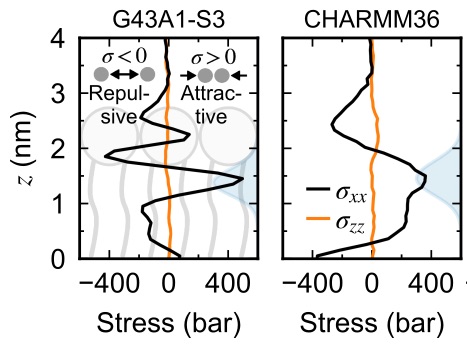 Winkeljohn, C. M., Himberg, B., and Vanegas, J.M. Balance of Solvent and Chain Interactions Determines the Local Stress State of Simulated Membranes. J. Phys. Chem. B 124, 32, 6963–6971 (2020) [PDF]
Abstract. Characterization of the internal mechanical state of model lipid membranes is essential to understand the microscopic underpinnings of biological functions such as membrane fission and organelle shaping within the context of elastic theories such as the Helfrich framework. Here, we compute lateral stress or pressure profiles from molecular dynamics simulations of lipid bilayers and water–vacuum interfaces to understand the role that solvent treatment and force-field parametrization plays on the local mechanical features of membranes[...]
Winkeljohn, C. M., Himberg, B., and Vanegas, J.M. Balance of Solvent and Chain Interactions Determines the Local Stress State of Simulated Membranes. J. Phys. Chem. B 124, 32, 6963–6971 (2020) [PDF]
Abstract. Characterization of the internal mechanical state of model lipid membranes is essential to understand the microscopic underpinnings of biological functions such as membrane fission and organelle shaping within the context of elastic theories such as the Helfrich framework. Here, we compute lateral stress or pressure profiles from molecular dynamics simulations of lipid bilayers and water–vacuum interfaces to understand the role that solvent treatment and force-field parametrization plays on the local mechanical features of membranes[...]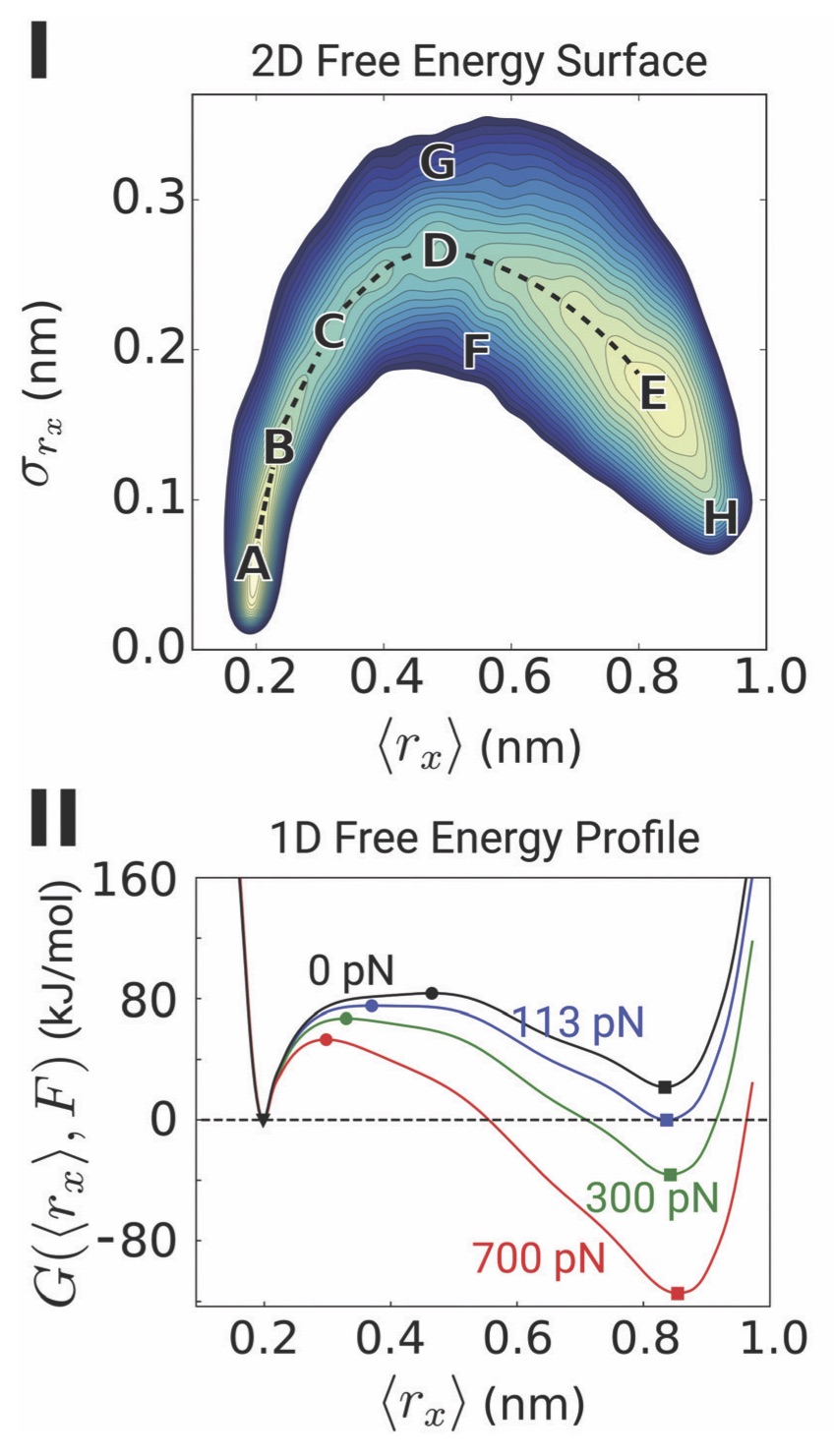 Torres-Sanchez, A., Vanegas, J. M., Purohit, P. K., and Arroyo, M. Combined molecular/continuum modeling reveals the role of friction during fast unfolding of coiled-coil proteins. Soft Matter 15, 4961-4975 (2019) [PDF]
Torres-Sanchez, A., Vanegas, J. M., Purohit, P. K., and Arroyo, M. Combined molecular/continuum modeling reveals the role of friction during fast unfolding of coiled-coil proteins. Soft Matter 15, 4961-4975 (2019) [PDF]Abstract. Coiled-coils are filamentous proteins that form the basic building block of important force-bearing cellular elements, such as intermediate filaments and myosin motors. In addition to their biological importance, coiled-coil proteins are increasingly used in new biomaterials including fibers, nanotubes, or hydrogels. Coiled-coils undergo a structural transition from an α-helical coil to an unfolded state upon extension, which allows them to sustain large strains and is critical for their biological function. [...]
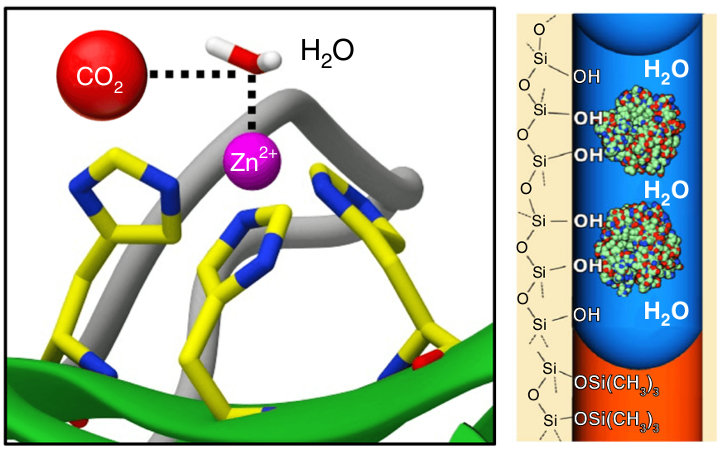 Fu, Y., Jiang, Y., Dunphy, D., Xiong, H., Coker, E., Chou, S., Zhang, H., Vanegas, J. M., Croissant, J., Cecchi, J., Rempe, S. B., and Brinker, C. J. Ultra-thin, enzymatic, nano-stabilized liquid membrane for CO2 separation. Nature Commun. 9, 990 (2018) [PDF]
Abstract. TThe limited flux and selectivities of current carbon dioxide membranes and the high costs associated with conventional absorption-based CO2 sequestration call for alternative CO2 separation approaches. Here we describe an enzymatically active, ultra-thin, biomimetic membrane enabling CO2 capture and separation under ambient pressure and temperature conditions. [...]
Fu, Y., Jiang, Y., Dunphy, D., Xiong, H., Coker, E., Chou, S., Zhang, H., Vanegas, J. M., Croissant, J., Cecchi, J., Rempe, S. B., and Brinker, C. J. Ultra-thin, enzymatic, nano-stabilized liquid membrane for CO2 separation. Nature Commun. 9, 990 (2018) [PDF]
Abstract. TThe limited flux and selectivities of current carbon dioxide membranes and the high costs associated with conventional absorption-based CO2 sequestration call for alternative CO2 separation approaches. Here we describe an enzymatically active, ultra-thin, biomimetic membrane enabling CO2 capture and separation under ambient pressure and temperature conditions. [...]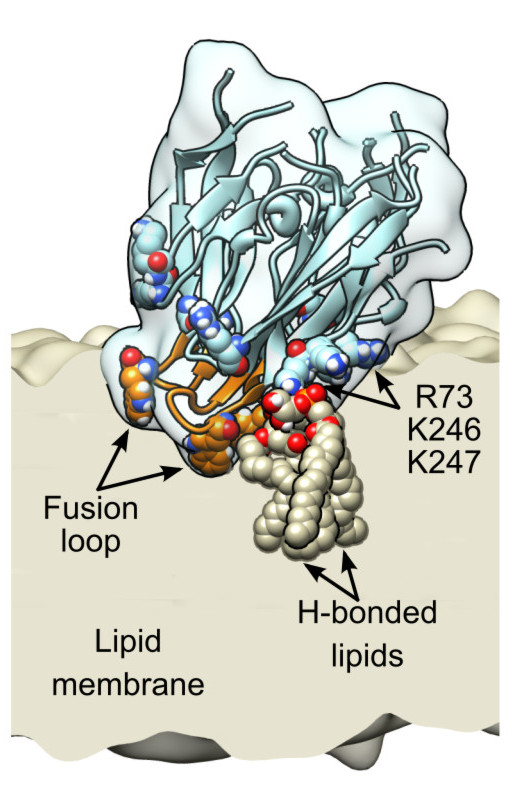 Vanegas, J. M., Heinrich, F., Rogers, D. M., Carson, B. D., La Bauve, S., Vernon, B. C., Akgun, B., Satija, S., Zheng, A., Kielian, M., Rempe, S. B., and Kent, M. S. Insertion of dengue E into lipid bilayers studied by neutron reflectivity and molecular dynamics simulations. Biochim. Biophys. Acta 1860, 1216 – 1230 (2018) [PDF]
Vanegas, J. M., Heinrich, F., Rogers, D. M., Carson, B. D., La Bauve, S., Vernon, B. C., Akgun, B., Satija, S., Zheng, A., Kielian, M., Rempe, S. B., and Kent, M. S. Insertion of dengue E into lipid bilayers studied by neutron reflectivity and molecular dynamics simulations. Biochim. Biophys. Acta 1860, 1216 – 1230 (2018) [PDF]Abstract. The envelope (E) protein of Dengue virus rearranges to a trimeric hairpin to mediate fusion of the viral and target membranes, which is essential for infectivity. Insertion of E into the target membrane serves to anchor E and possibly also to disrupt local order within the membrane[...]
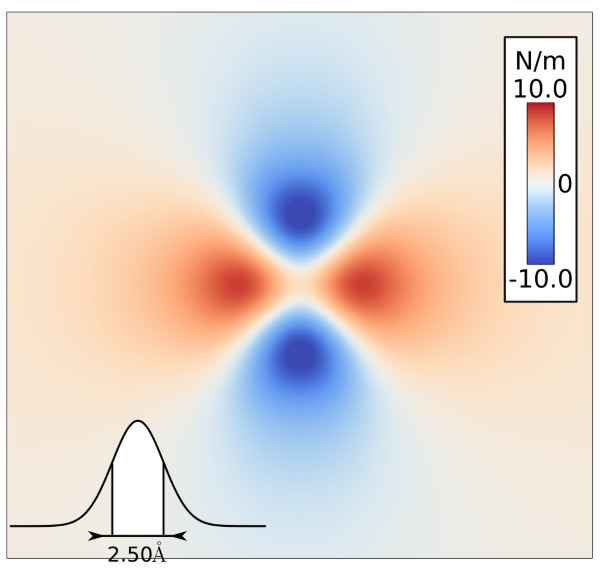
 Torres-Sanchez, A., Vanegas, J. M., and Arroyo, M. Geometric derivation of the microscopic stress: A covariant central force decomposition. J. Mech. Phys. Solids 93, 224 – 239 (2016) [PDF]
Torres-Sanchez, A., Vanegas, J. M., and Arroyo, M. Geometric derivation of the microscopic stress: A covariant central force decomposition. J. Mech. Phys. Solids 93, 224 – 239 (2016) [PDF]Abstract. We revisit the derivation of the microscopic stress, linking the statistical mechanics of particle systems and continuum mechanics. The starting point in our geometric derivation is the Doyle–Ericksen formula [...]
 Torres-Sanchez, A., Vanegas, J. M., and Arroyo, M. Examining the mechanical equilibrium of microscopic stresses in molecular simulations. Phys. Rev. Lett. 114, 258102 (2015). [PDF]
Torres-Sanchez, A., Vanegas, J. M., and Arroyo, M. Examining the mechanical equilibrium of microscopic stresses in molecular simulations. Phys. Rev. Lett. 114, 258102 (2015). [PDF]Abstract. The microscopic stress field provides a unique connection between atomistic simulations and mechanics at the nanoscale. However, its definition remains ambiguous. Rather than a mere theoretical [...]
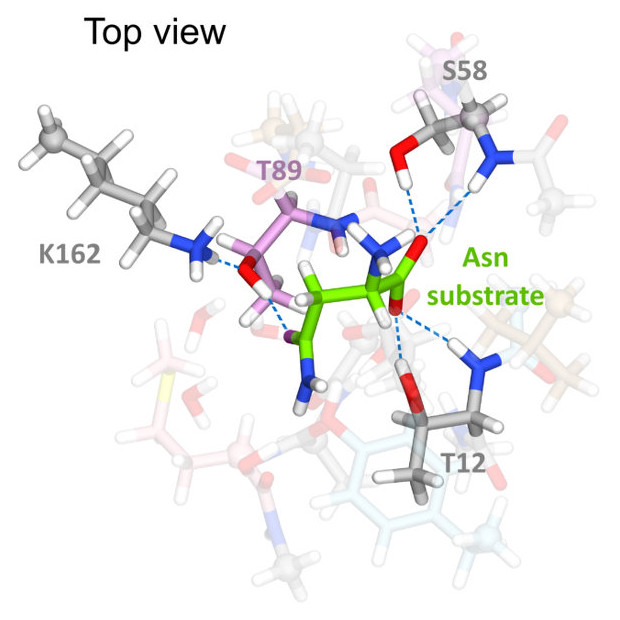 Anishkin, A., Vanegas, J. M., Rogers, D. M., Lorenzi, P. L., Chan, W. K., Purwaha, P., Weinstein, J. N., Sukharev, S., and Rempe, S. B. Catalytic role of the substrate defines specificity of therapeutic L-asparaginase. J. Mol. Biol. 427, (17), 2867–2885 (2015) [PDF]
Anishkin, A., Vanegas, J. M., Rogers, D. M., Lorenzi, P. L., Chan, W. K., Purwaha, P., Weinstein, J. N., Sukharev, S., and Rempe, S. B. Catalytic role of the substrate defines specificity of therapeutic L-asparaginase. J. Mol. Biol. 427, (17), 2867–2885 (2015) [PDF]Abstract. Type II bacterial l-asparaginases (l-ASP) have played an important therapeutic role in cancer treatment for over four decades, yet their exact [...]
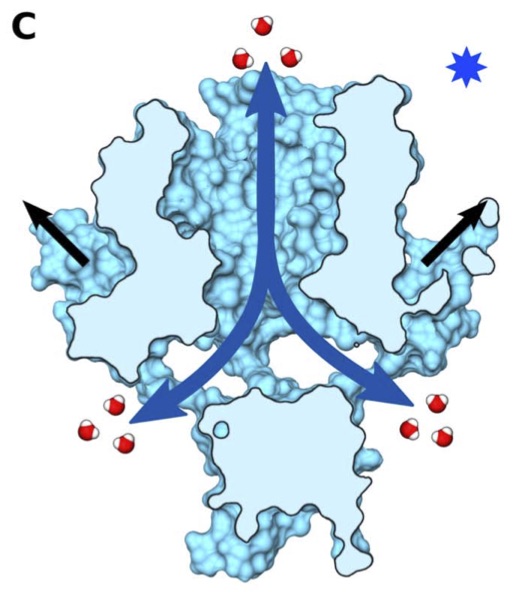 Vanegas, J. M. and Arroyo, M. Force transduction and lipid binding in MscL: A continuum-molecular approach. PLoS ONE, 9 (12), e113947. (2014) [PDF]
Vanegas, J. M. and Arroyo, M. Force transduction and lipid binding in MscL: A continuum-molecular approach. PLoS ONE, 9 (12), e113947. (2014) [PDF]Abstract. The bacterial mechanosensitive channel MscL, a small protein mainly activated by membrane tension, is a central model system to study the transduction of mechanical stimuli into chemical signals. Mutagenic studies suggest that MscL gating strongly depends on both intra-protein and interfacial lipid-protein interactions. However, there is a gap between this [...]
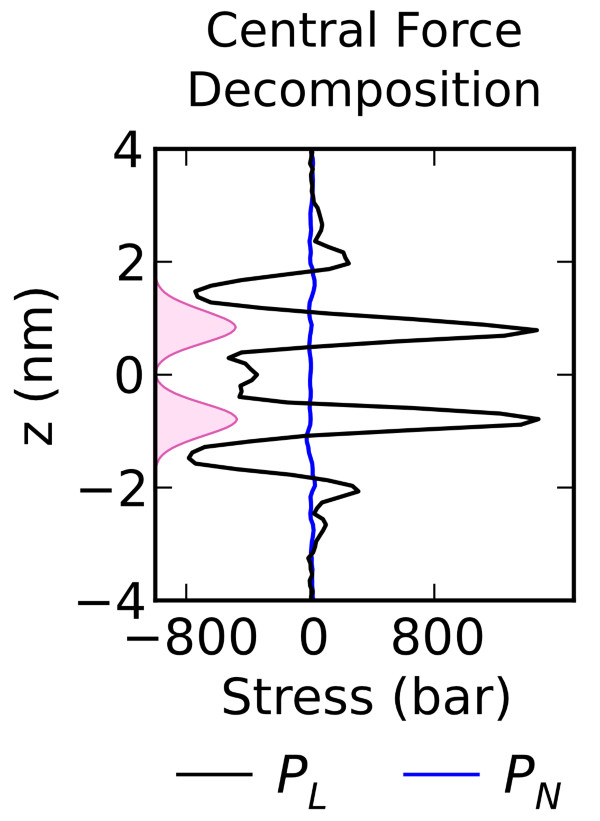 Vanegas, J. M., Torres-Sanchez, A., and Arroyo, M. Importance of force decomposition for local stress calculations in biomembrane molecular simulations. J. Chem. Theory Comput., 10, 691-702. (2014) [PDF]
Vanegas, J. M., Torres-Sanchez, A., and Arroyo, M. Importance of force decomposition for local stress calculations in biomembrane molecular simulations. J. Chem. Theory Comput., 10, 691-702. (2014) [PDF]Abstract. Local stress fields are routinely computed from molecular dynamics trajectories to understand the structure and mechanical properties of lipid bilayers. These calculations can be systematically understood with the Irving–Kirkwood–Noll theory. In identifying the stress tensor, a crucial step is the decomposition of the forces on the particles into pairwise [...]